DeepTracer™
Technology No. 49069,49151,8133
A software tool for ultra-fast, fully automated, and accurate de novo protein large complex structure prediction from 3D electron microscopy map. DeepTracer uses deep learning methods and optimization techniques to predict the locations of the atoms, secondary structure elements, and amino acid types. It targets the ab initio all-atom protein structure prediction problem for extremely large complexes, such as viruses, bacteria, and other molecular machines. The work has been featured on PNAS research highlights and Nature Computational Science research highlights.
For more information: https://deeptracer.uw.edu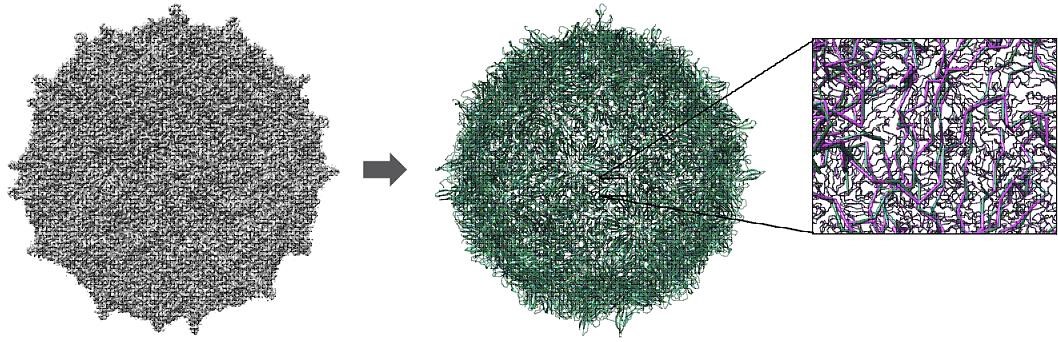
International Requests:
University of Washington agreements require additional review if the requesting entity is located in, or affiliated with the government of, China (Including Hong Kong; not including Taiwan), Iran, North Korea, Russia, or Syria. For requests from these countries, please allow for an additional month of processing time for a response.Available Licenses
Academic License:See right panel to license this product.
-
expand_more library_books References (2)
- Jonas Pfab, Nhut Minh Phan, and Dong Si , DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes
- Fernando Chirigati , Predicting protein structure from cryo-EM data
-
expand_more cloud_download Supporting documents (0)Additional files may be available once you've completed the transaction for this product. If you've already done so, please log into your account and visit My account / Downloads section to view them.